Cromwell¶
This workflow engine is focused on the genomics community. Originally developed by the Broad Institute, Cromwell is also used in the GATK Best Practices genome analysis pipeline.
Logo |
|
|---|---|
Website |
|
Repository |
|
Byline |
Scientific workflow engine designed for simplicity & scalability. Trivially transition between one off use cases to massive scale production environments |
License |
BSD 3-clause |
Project age |
7 years 7 months |
Backers |
|
Size score (1 to 10, higher is better) |
5.25 |
Trend score (1 to 10, higher is better) |
4.25 |
Education Resources¶
URL |
Resource Type |
Description |
|---|---|---|
Documentation |
Official project documentation. |
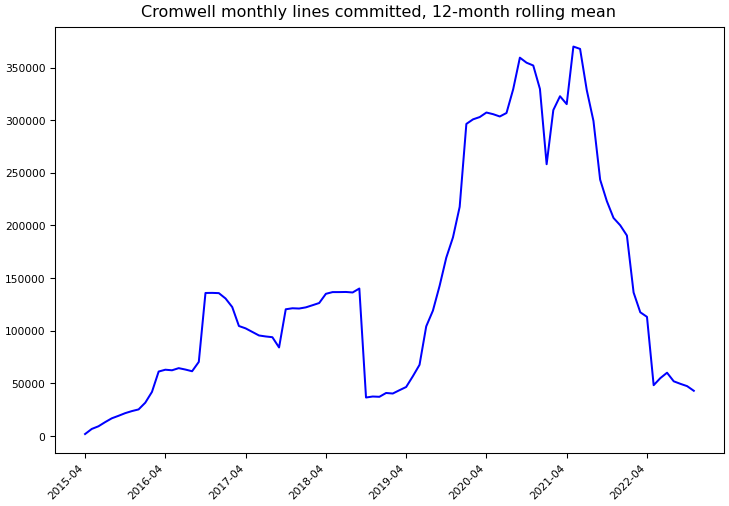
Git Commit Statistics¶
Statistics computed using Git data through November 30, 2022.
Statistic |
Lifetime |
Last 12 Months |
|---|---|---|
Commits |
43,398 |
2,039 |
Lines committed |
13,303,244 |
513,806 |
Unique committers |
151 |
20 |
Core committers |
14 |
11 |
Similar Projects¶
Project |
Size Score |
Trend Score |
Byline |
|---|---|---|---|
8.25 |
8.5 |
Apache Airflow - A platform to programmatically author, schedule, and monitor workflows |
|
5.0 |
3.75 |
Luigi is a Python module that helps you build complex pipelines of batch jobs. It handles dependency resolution, workflow management, visualization etc. It also comes with Hadoop support built in. |
|
7.5 |
6.5 |
The Snakemake workflow management system is a tool to create reproducible and scalable data analyses |
